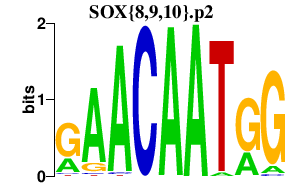
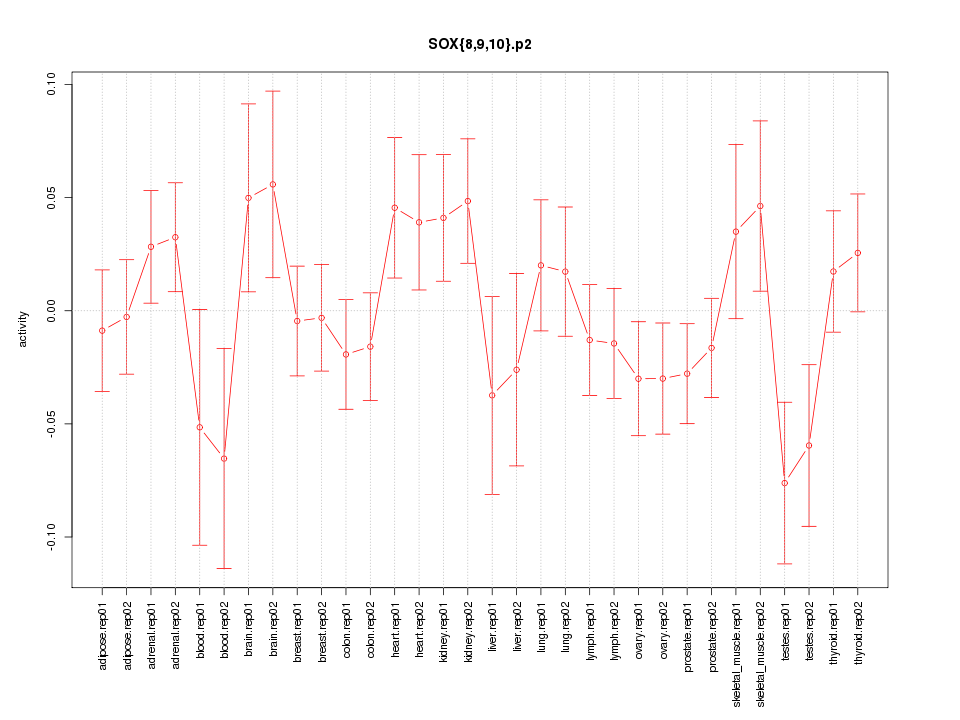
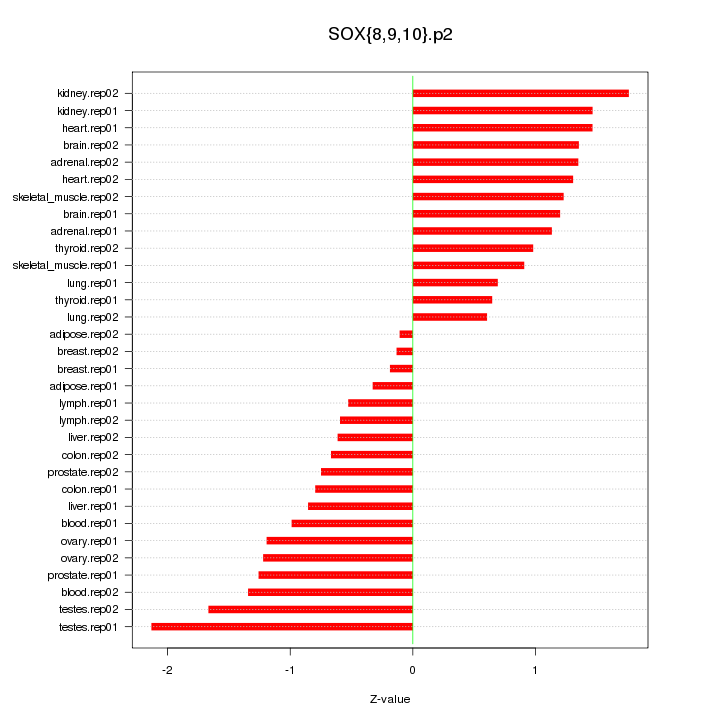
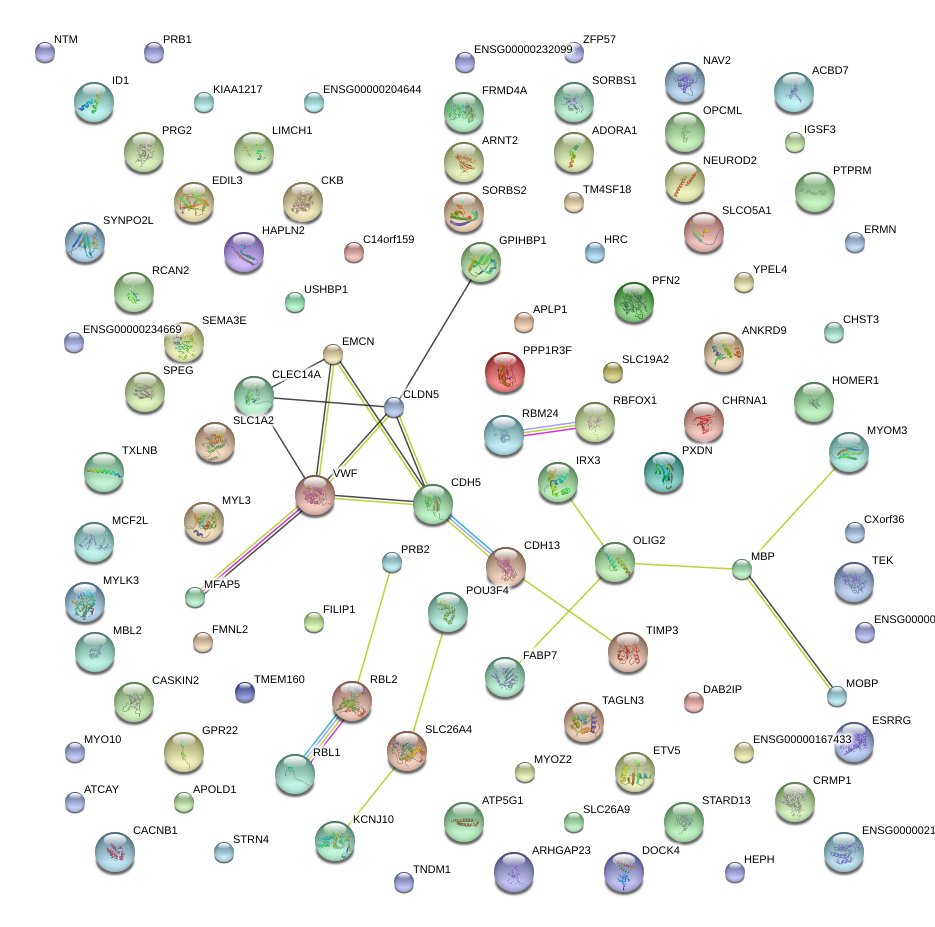
|
chr3_-_46904879
|
4.632
|
NM_000258
|
MYL3
|
myosin, light chain 3, alkali; ventricular, skeletal, slow
|
|
chr1_+_156589085
|
3.228
|
NM_021817
|
HAPLN2
|
hyaluronan and proteoglycan link protein 2
|
|
chr2_-_158182154
|
2.879
|
NM_020711
|
ERMN
|
ermin, ERM-like protein
|
|
chr3_-_185826748
|
2.490
|
NM_004454
|
ETV5
|
ets variant 5
|
|
chr19_-_49658645
|
2.417
|
NM_002152
|
HRC
|
histidine rich calcium binding protein
|
|
chr6_-_46293215
|
2.341
|
|
RCAN2
|
regulator of calcineurin 2
|
|
chrX_+_49126326
|
2.148
|
|
PPP1R3F
|
protein phosphatase 1, regulatory subunit 3F
|
|
chr1_-_205912546
|
2.090
|
NM_052934
NM_134325
|
SLC26A9
|
solute carrier family 26, member 9
|
|
chr6_-_46293404
|
1.925
|
|
RCAN2
|
regulator of calcineurin 2
|
|
chr14_-_102975996
|
1.871
|
|
ANKRD9
|
ankyrin repeat domain 9
|
|
chr8_-_70745403
|
1.849
|
NM_001146009
|
SLCO5A1
|
solute carrier organic anion transporter family, member 5A1
|
|
chr10_-_15130773
|
1.723
|
NM_001039844
|
ACBD7
|
acyl-CoA binding domain containing 7
|
|
chr4_-_186732047
|
1.706
|
NM_001145672
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr11_-_35440514
|
1.697
|
|
SLC1A2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2
|
|
chr6_-_139613114
|
1.671
|
NM_153235
|
TXLNB
|
taxilin beta
|
|
chr14_-_102976096
|
1.645
|
NM_152326
|
ANKRD9
|
ankyrin repeat domain 9
|
|
chr10_-_97321094
|
1.590
|
NM_015385
NM_024991
|
SORBS1
|
sorbin and SH3 domain containing 1
|
|
chr4_+_41614911
|
1.556
|
NM_001112719
NM_001112720
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr8_+_144295067
|
1.514
|
NM_178172
|
GPIHBP1
|
glycosylphosphatidylinositol anchored high density lipoprotein binding protein 1
|
|
chr5_-_83680191
|
1.504
|
|
EDIL3
|
EGF-like repeats and discoidin I-like domains 3
|
|
chr16_+_6069094
|
1.464
|
NM_001142333
NM_018723
|
RBFOX1
|
RNA binding protein, fox-1 homolog (C. elegans) 1
|
|
chr16_+_82660641
|
1.463
|
|
CDH13
|
cadherin 13, H-cadherin (heart)
|
|
chr19_-_47551881
|
1.447
|
NM_017854
|
TMEM160
|
transmembrane protein 160
|
|
chr16_-_46782104
|
1.430
|
NM_182493
|
MYLK3
|
myosin light chain kinase 3
|
|
chr1_-_160039946
|
1.422
|
NM_002241
|
KCNJ10
|
potassium inwardly-rectifying channel, subfamily J, member 10
|
|
chr4_+_41614933
|
1.404
|
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr4_-_186732208
|
1.394
|
NM_001145671
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr1_-_117210301
|
1.387
|
NM_001007237
NM_001542
|
IGSF3
|
immunoglobulin superfamily, member 3
|
|
chr2_-_175629140
|
1.369
|
NM_000079
NM_001039523
|
CHRNA1
|
cholinergic receptor, nicotinic, alpha 1 (muscle)
|
|
chr9_+_27109138
|
1.368
|
NM_000459
|
TEK
|
TEK tyrosine kinase, endothelial
|
|
chr14_-_103989112
|
1.356
|
|
CKB
|
creatine kinase, brain
|
|
chr16_+_82660573
|
1.354
|
|
CDH13
|
cadherin 13, H-cadherin (heart)
|
|
chr5_-_83680590
|
1.350
|
NM_005711
|
EDIL3
|
EGF-like repeats and discoidin I-like domains 3
|
|
chr12_-_8815266
|
1.349
|
|
MFAP5
|
microfibrillar associated protein 5
|
|
chr19_+_3880617
|
1.344
|
NM_033064
|
ATCAY
|
ataxia, cerebellar, Cayman type
|
|
chr12_-_6233684
|
1.298
|
NM_000552
|
VWF
|
von Willebrand factor
|
|
chr5_-_78809658
|
1.297
|
NM_004272
|
HOMER1
|
homer homolog 1 (Drosophila)
|
|
chr21_+_34398210
|
1.291
|
NM_005806
|
OLIG2
|
oligodendrocyte lineage transcription factor 2
|
|
chr10_-_75410778
|
1.258
|
NM_024875
|
SYNPO2L
|
synaptopodin 2-like
|
|
chr4_+_41614725
|
1.254
|
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr7_+_107301079
|
1.250
|
NM_000441
|
SLC26A4
|
solute carrier family 26, member 4
|
|
chr17_-_37764165
|
1.247
|
NM_006160
|
NEUROD2
|
neurogenic differentiation 2
|
|
chr6_-_29644930
|
1.246
|
NM_001109809
|
ZFP57
|
zinc finger protein 57 homolog (mouse)
|
|
chr14_-_38725539
|
1.244
|
NM_175060
|
CLEC14A
|
C-type lectin domain family 14, member A
|
|
chr19_+_36359346
|
1.241
|
NM_001024807
NM_005166
|
APLP1
|
amyloid beta (A4) precursor-like protein 1
|
|
chr22_-_19512859
|
1.239
|
NM_001130861
NM_003277
|
CLDN5
|
claudin 5
|
|
chr16_+_82660390
|
1.234
|
NM_001220488
NM_001220489
NM_001220490
NM_001220491
NM_001220492
NM_001257
|
CDH13
|
cadherin 13, H-cadherin (heart)
|
|
chr1_-_24438625
|
1.233
|
NM_152372
|
MYOM3
|
myomesin family, member 3
|
|
chrX_-_45060081
|
1.221
|
NM_024689
NM_176819
|
CXorf36
|
chromosome X open reading frame 36
|
|
chr11_+_20049128
|
1.209
|
|
NAV2
|
neuron navigator 2
|
|
chr8_-_70745624
|
1.183
|
NM_001146008
|
SLCO5A1
|
solute carrier organic anion transporter family, member 5A1
|
|
chr15_+_80696569
|
1.172
|
NM_014862
|
ARNT2
|
aryl-hydrocarbon receptor nuclear translocator 2
|
|
chr18_+_7946885
|
1.171
|
|
PTPRM
|
protein tyrosine phosphatase, receptor type, M
|
|
chr17_-_37353731
|
1.171
|
NM_000723
NM_199247
NM_199248
|
CACNB1
|
calcium channel, voltage-dependent, beta 1 subunit
|
|
chr7_+_107110501
|
1.170
|
NM_005295
|
GPR22
|
G protein-coupled receptor 22
|
|
chr14_-_103989195
|
1.158
|
NM_001823
|
CKB
|
creatine kinase, brain
|
|
chrX_+_49126299
|
1.149
|
NM_001184745
NM_033215
|
PPP1R3F
|
protein phosphatase 1, regulatory subunit 3F
|
|
chr4_+_120056938
|
1.136
|
NM_016599
|
MYOZ2
|
myozenin 2
|
|
chr3_-_149051258
|
1.136
|
NM_001184723
NM_138786
|
TM4SF18
|
transmembrane 4 L six family member 18
|
|
chr4_-_101439071
|
1.126
|
NM_001159694
NM_016242
|
EMCN
|
endomucin
|
|
chr13_+_113656076
|
1.101
|
|
MCF2L
|
MCF.2 cell line derived transforming sequence-like
|
|
chr20_+_30193083
|
1.100
|
NM_002165
NM_181353
|
ID1
|
inhibitor of DNA binding 1, dominant negative helix-loop-helix protein
|
|
chr12_-_11548485
|
1.097
|
NM_006248
|
PRB1
PRB2
|
proline-rich protein BstNI subfamily 1
proline-rich protein BstNI subfamily 2
|
|
chr7_-_83270746
|
1.096
|
NM_001178129
|
SEMA3E
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3E
|
|
chr10_+_24498088
|
1.093
|
|
KIAA1217
|
KIAA1217
|
|
chrX_+_82763268
|
1.093
|
NM_000307
|
POU3F4
|
POU class 3 homeobox 4
|
|
chr7_-_111846192
|
1.091
|
|
DOCK4
|
dedicator of cytokinesis 4
|
|
chr4_+_41540320
|
1.085
|
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr17_-_73505914
|
1.081
|
NM_001142643
|
CASKIN2
|
CASK interacting protein 2
|
|
chr19_-_17375522
|
1.072
|
NM_031941
|
USHBP1
|
Usher syndrome 1C binding protein 1
|
|
chr14_+_91580713
|
1.072
|
|
C14orf159
|
chromosome 14 open reading frame 159
|
|
chr22_+_33197678
|
1.064
|
|
TIMP3
|
TIMP metallopeptidase inhibitor 3
|
|
chr14_-_38725253
|
1.060
|
|
CLEC14A
|
C-type lectin domain family 14, member A
|
|
chr11_+_131781303
|
1.056
|
|
NTM
|
neurotrimin
|
|
chr11_-_57417247
|
1.054
|
NM_145008
|
YPEL4
|
yippee-like 4 (Drosophila)
|
|
chr1_-_217311095
|
1.052
|
NM_001134285
|
ESRRG
|
estrogen-related receptor gamma
|
|
chr18_-_74729049
|
1.050
|
NM_001025081
NM_001025090
NM_001025092
NM_002385
|
MBP
|
myelin basic protein
|
|
chr10_-_13749622
|
1.033
|
|
FRMD4A
|
FERM domain containing 4A
|
|
chr6_+_17282931
|
1.032
|
NM_153020
|
RBM24
|
RNA binding motif protein 24
|
|
chr17_+_46970128
|
1.032
|
NM_001002027
NM_005175
|
ATP5G1
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C1 (subunit 9)
|
|
chr12_-_11508509
|
1.030
|
NM_005039
NM_199353
NM_199354
|
PRB1
|
proline-rich protein BstNI subfamily 1
|
|
chr12_+_12938540
|
1.029
|
NM_030817
|
APOLD1
|
apolipoprotein L domain containing 1
|
|
chr16_-_54319966
|
1.028
|
|
IRX3
|
iroquois homeobox 3
|
|
chr3_-_149688886
|
1.026
|
|
PFN2
|
profilin 2
|
|
chr2_-_56412904
|
1.013
|
|
LOC100129434
|
uncharacterized LOC100129434
|
|
chr5_-_16936371
|
1.009
|
NM_012334
|
MYO10
|
myosin X
|
|
chr5_+_140739589
|
1.008
|
NM_018923
NM_032096
|
PCDHGB2
|
protocadherin gamma subfamily B, 2
|
|
chr16_+_66400524
|
1.003
|
NM_001795
|
CDH5
|
cadherin 5, type 2 (vascular endothelium)
|
|
chr10_+_73723992
|
1.003
|
NM_004273
|
CHST3
|
carbohydrate (chondroitin 6) sulfotransferase 3
|
|
chr16_+_66400585
|
0.988
|
|
CDH5
|
cadherin 5, type 2 (vascular endothelium)
|
|
chr9_+_124461365
|
0.974
|
|
DAB2IP
|
DAB2 interacting protein
|
|
chr3_+_111717585
|
0.970
|
NM_001008272
NM_013259
|
TAGLN3
|
transgelin 3
|
|
chr19_+_3880675
|
0.965
|
|
ATCAY
|
ataxia, cerebellar, Cayman type
|
|
chr19_+_3880660
|
0.963
|
|
ATCAY
|
ataxia, cerebellar, Cayman type
|
|
chr6_-_76203256
|
0.956
|
NM_015687
|
FILIP1
|
filamin A interacting protein 1
|
|
chr22_+_33197744
|
0.955
|
|
TIMP3
|
TIMP metallopeptidase inhibitor 3
|
|
chr3_+_147111221
|
0.953
|
|
|
|
|
chr1_+_203096835
|
0.950
|
NM_000674
NM_001048230
|
ADORA1
|
adenosine A1 receptor
|
|
chr3_+_181417387
|
0.946
|
|
SOX2-OT
|
SOX2 overlapping transcript (non-protein coding)
|
|
chrX_+_49126480
|
0.944
|
|
PPP1R3F
|
protein phosphatase 1, regulatory subunit 3F
|
|
chr3_+_39509069
|
0.942
|
NM_182935
|
MOBP
|
myelin-associated oligodendrocyte basic protein
|
|
chr6_-_72130447
|
0.930
|
|
LINC00472
|
long intergenic non-protein coding RNA 472
|
|
chr18_-_74728963
|
0.927
|
|
MBP
|
myelin basic protein
|
|
chr14_+_91580467
|
0.924
|
|
C14orf159
|
chromosome 14 open reading frame 159
|
|
chr13_-_33760215
|
0.923
|
NM_001243466
NM_178007
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr12_-_8815370
|
0.922
|
NM_003480
|
MFAP5
|
microfibrillar associated protein 5
|
|
chr17_+_40119669
|
0.922
|
|
|
|
|
chr10_+_24497719
|
0.921
|
NM_001098501
NM_019590
|
KIAA1217
|
KIAA1217
|
|
chr3_+_111718006
|
0.909
|
NM_001008273
|
TAGLN3
|
transgelin 3
|
|
chr6_+_123100860
|
0.905
|
|
FABP7
|
fatty acid binding protein 7, brain
|
|
chr14_+_91580967
|
0.895
|
NM_001102367
|
C14orf159
|
chromosome 14 open reading frame 159
|
|
chr4_-_5890142
|
0.893
|
|
CRMP1
|
collapsin response mediator protein 1
|
|
chr1_-_169455099
|
0.887
|
NM_006996
|
SLC19A2
|
solute carrier family 19 (thiamine transporter), member 2
|
|
chr2_+_220309373
|
0.886
|
|
SPEG
|
SPEG complex locus
|
|
chr17_+_36584692
|
0.883
|
NM_001199417
|
ARHGAP23
|
Rho GTPase activating protein 23
|
|
chrX_+_65384078
|
0.881
|
NM_001130860
NM_014799
|
HEPH
|
hephaestin
|
|
chr2_+_153191830
|
0.878
|
|
FMNL2
|
formin-like 2
|
|
chr7_-_131241321
|
0.868
|
NM_001018111
NM_005397
|
PODXL
|
podocalyxin-like
|
|
chr15_+_71184781
|
0.866
|
NM_001199017
NM_017691
NM_001199018
|
LRRC49
|
leucine rich repeat containing 49
|
|
chr12_+_53443834
|
0.865
|
NM_170754
|
TENC1
|
tensin like C1 domain containing phosphatase (tensin 2)
|
|
chr6_+_17282294
|
0.860
|
NM_001143941
|
RBM24
|
RNA binding motif protein 24
|
|
chr9_+_2622134
|
0.860
|
|
VLDLR
|
very low density lipoprotein receptor
|
|
chr11_+_131780537
|
0.857
|
NM_001144058
NM_001144059
NM_016522
|
NTM
|
neurotrimin
|
|
chr16_+_66400559
|
0.856
|
|
CDH5
|
cadherin 5, type 2 (vascular endothelium)
|
|
chr21_+_41239346
|
0.855
|
NM_006198
|
PCP4
|
Purkinje cell protein 4
|
|
chr12_-_18243052
|
0.855
|
NM_024730
|
RERGL
|
RERG/RAS-like
|
|
chr7_-_44887663
|
0.854
|
|
H2AFV
|
H2A histone family, member V
|
|
chr2_+_198669316
|
0.854
|
NM_006226
|
PLCL1
|
phospholipase C-like 1
|
|
chr8_+_21924369
|
0.853
|
|
EPB49
|
erythrocyte membrane protein band 4.9 (dematin)
|
|
chr17_+_8213555
|
0.853
|
NM_173728
|
ARHGEF15
|
Rho guanine nucleotide exchange factor (GEF) 15
|
|
chr15_+_82944821
|
0.842
|
|
AGSK1
|
golgin subfamily A member 2-like
|
|
chr10_+_24755459
|
0.842
|
|
KIAA1217
|
KIAA1217
|
|
chr15_+_82944772
|
0.834
|
|
AGSK1
|
golgin subfamily A member 2-like
|
|
chr2_+_159313407
|
0.834
|
|
PKP4
|
plakophilin 4
|
|
chr17_-_38210611
|
0.832
|
|
MED24
|
mediator complex subunit 24
|
|
chr12_-_11002051
|
0.832
|
NM_001098538
NM_007244
|
PRR4
|
proline rich 4 (lacrimal)
|
|
chr1_-_27701302
|
0.832
|
NM_003665
NM_173452
|
FCN3
|
ficolin (collagen/fibrinogen domain containing) 3 (Hakata antigen)
|
|
chr19_-_55660557
|
0.830
|
NM_001126132
NM_001126133
NM_003283
|
TNNT1
|
troponin T type 1 (skeletal, slow)
|
|
chr6_-_110736748
|
0.823
|
NM_003649
NM_004032
|
DDO
|
D-aspartate oxidase
|
|
chr1_+_26503973
|
0.818
|
NM_006314
|
CNKSR1
|
connector enhancer of kinase suppressor of Ras 1
|
|
chr10_+_6244839
|
0.817
|
NM_004566
|
PFKFB3
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3
|
|
chr8_+_40010986
|
0.814
|
NM_020130
|
C8orf4
|
chromosome 8 open reading frame 4
|
|
chr5_+_140593508
|
0.805
|
NM_018933
|
PCDHB13
|
protocadherin beta 13
|
|
chr1_-_85930610
|
0.803
|
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1
|
|
chr4_-_177190258
|
0.798
|
NM_080874
|
ASB5
|
ankyrin repeat and SOCS box containing 5
|
|
chr20_+_58179601
|
0.797
|
NM_080672
|
PHACTR3
|
phosphatase and actin regulator 3
|
|
chr5_-_41510627
|
0.794
|
NM_001005473
|
PLCXD3
|
phosphatidylinositol-specific phospholipase C, X domain containing 3
|
|
chr1_-_149858114
|
0.790
|
NM_003528
|
HIST2H2BE
|
histone cluster 2, H2be
|
|
chr1_-_186417855
|
0.789
|
NM_022576
|
PDC
|
phosducin
|
|
chr1_-_79472360
|
0.789
|
NM_022159
|
ELTD1
|
EGF, latrophilin and seven transmembrane domain containing 1
|
|
chr15_-_83876769
|
0.785
|
|
HDGFRP3
|
hepatoma-derived growth factor, related protein 3
|
|
chr2_-_45236521
|
0.782
|
NM_016932
|
SIX2
|
SIX homeobox 2
|
|
chr14_+_91580356
|
0.781
|
NM_001102366
NM_001102368
NM_001102369
NM_024952
|
C14orf159
|
chromosome 14 open reading frame 159
|
|
chr17_-_55980749
|
0.780
|
NM_017949
|
CUEDC1
|
CUE domain containing 1
|
|
chr9_+_2622099
|
0.780
|
|
VLDLR
|
very low density lipoprotein receptor
|
|
chr5_-_146833150
|
0.778
|
NM_001387
|
DPYSL3
|
dihydropyrimidinase-like 3
|
|
chr3_-_62860561
|
0.778
|
|
|
|
|
chr5_+_140602914
|
0.774
|
NM_018934
|
PCDHB14
|
protocadherin beta 14
|
|
chr14_-_94442894
|
0.774
|
|
ASB2
|
ankyrin repeat and SOCS box containing 2
|
|
chr9_+_132934685
|
0.773
|
NM_014286
|
NCS1
|
neuronal calcium sensor 1
|
|
chr9_-_14398981
|
0.772
|
NM_001190738
|
NFIB
|
nuclear factor I/B
|
|
chr6_+_121756744
|
0.768
|
NM_000165
|
GJA1
|
gap junction protein, alpha 1, 43kDa
|
|
chr20_+_61147924
|
0.767
|
|
C20orf166
|
chromosome 20 open reading frame 166
|
|
chr2_-_175869910
|
0.764
|
NM_001822
NM_001025201
|
CHN1
|
chimerin (chimaerin) 1
|
|
chr4_+_115543522
|
0.762
|
NM_003360
|
UGT8
|
UDP glycosyltransferase 8
|
|
chrY_-_21239250
|
0.760
|
|
TTTY14
|
testis-specific transcript, Y-linked 14 (non-protein coding)
|
|
chr16_+_2083264
|
0.760
|
NM_001252073
NM_001252075
NM_001252076
|
SLC9A3R2
|
solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 2
|
|
chr4_+_184827882
|
0.757
|
|
|
|
|
chr5_+_140797210
|
0.755
|
NM_032101
|
PCDHGB7
|
protocadherin gamma subfamily B, 7
|
|
chr6_+_123100645
|
0.748
|
NM_001446
|
FABP7
|
fatty acid binding protein 7, brain
|
|
chrX_+_152954965
|
0.745
|
NM_001142806
|
SLC6A8
|
solute carrier family 6 (neurotransmitter transporter, creatine), member 8
|
|
chr4_+_184827805
|
0.745
|
|
STOX2
|
storkhead box 2
|
|
chr2_-_161056762
|
0.744
|
|
ITGB6
|
integrin, beta 6
|
|
chr10_-_98119058
|
0.736
|
NM_001040102
NM_001040103
NM_033207
|
OPALIN
|
oligodendrocytic myelin paranodal and inner loop protein
|
|
chr14_+_63671101
|
0.721
|
NM_020663
|
RHOJ
|
ras homolog gene family, member J
|
|
chr12_-_88974094
|
0.720
|
NM_000899
NM_003994
|
KITLG
|
KIT ligand
|
|
chr16_+_58534045
|
0.716
|
NM_001242833
NM_001242834
NM_001242835
NM_001242836
|
NDRG4
|
NDRG family member 4
|
|
chr8_-_82395401
|
0.711
|
NM_001442
|
FABP4
|
fatty acid binding protein 4, adipocyte
|
|
chr2_-_151344145
|
0.708
|
NM_005168
|
RND3
|
Rho family GTPase 3
|
|
chr18_+_55711388
|
0.705
|
NM_001144967
NM_001243960
NM_015277
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr14_+_55168831
|
0.702
|
|
SAMD4A
|
sterile alpha motif domain containing 4A
|
|
chrX_-_13835313
|
0.699
|
NM_001001995
NM_001001996
NM_005278
|
GPM6B
|
glycoprotein M6B
|
|
chr12_-_71003622
|
0.694
|
NM_001206971
NM_001206972
NM_002837
|
PTPRB
|
protein tyrosine phosphatase, receptor type, B
|
|
chr7_-_111846384
|
0.689
|
NM_014705
|
DOCK4
|
dedicator of cytokinesis 4
|
|
chr1_+_47489239
|
0.688
|
NM_178033
|
CYP4X1
|
cytochrome P450, family 4, subfamily X, polypeptide 1
|
|
chr4_+_41540187
|
0.688
|
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr5_+_149980627
|
0.687
|
NM_001166208
|
SYNPO
|
synaptopodin
|
|
chr17_+_44928947
|
0.687
|
NM_003396
|
WNT9B
|
wingless-type MMTV integration site family, member 9B
|
|
chr16_-_75285383
|
0.685
|
NM_001170717
NM_014567
|
BCAR1
|
breast cancer anti-estrogen resistance 1
|
|
chr5_+_180467224
|
0.683
|
NM_152547
|
BTNL9
|
butyrophilin-like 9
|
|
chr6_-_11232900
|
0.683
|
|
NEDD9
|
neural precursor cell expressed, developmentally down-regulated 9
|
|
chr4_+_183245136
|
0.682
|
NM_001080477
|
ODZ3
|
odz, odd Oz/ten-m homolog 3 (Drosophila)
|
|
chr2_-_170218972
|
0.680
|
|
LRP2
|
low density lipoprotein receptor-related protein 2
|
|
chr3_-_9439122
|
0.679
|
|
LOC440944
|
uncharacterized LOC440944
|
|
chr2_-_55277672
|
0.678
|
|
RTN4
|
reticulon 4
|
|
chr10_+_70748476
|
0.677
|
NM_015634
|
KIAA1279
|
KIAA1279
|
|
chr14_-_93214873
|
0.675
|
NM_001008530
NM_005606
|
LGMN
|
legumain
|
|
chr15_-_93277303
|
0.674
|
|
FAM174B
|
family with sequence similarity 174, member B
|
|
chr7_-_117513466
|
0.671
|
|
CTTNBP2
|
cortactin binding protein 2
|
|
chr22_-_36013303
|
0.671
|
NM_005368
NM_203378
|
MB
|
myoglobin
|